Sampling From Unknown Distributions¶
Feng Li
School of Statistics and Mathematics
Central University of Finance and Economics
Direct Methods¶
Only need random numbers from uniform
Assume that we have a way to simulate from a uniform distribution between $0$ and $1$, $u\sim U(0,1)$
If this is available, it is possible to simulate many other probability distributions.
The most simple method is the Direct Method
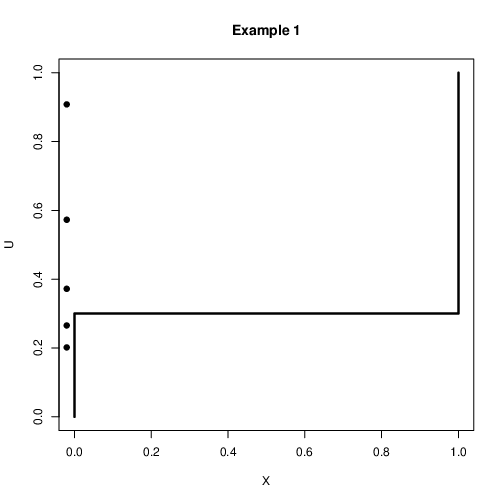
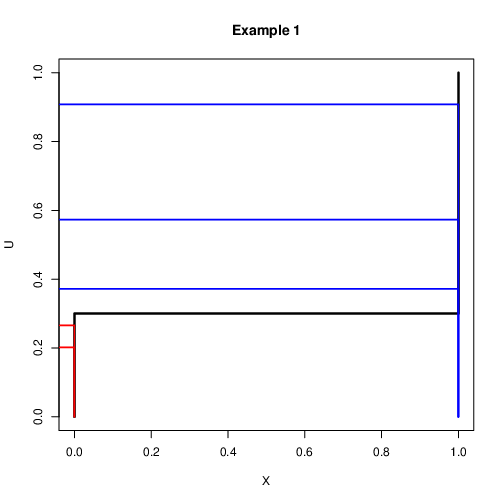
Discrete Case: Example 1¶
Assume that we want to simulate a binary variable $X$ with $\mbox{Pr}(X=1)=0.3$ and $\mbox{Pr}(X=0)=0.7$
Let $u\sim U(0,1)$. Then the following rule can be used $$x=\left{\begin{array}{c}
1 \quad \mbox{if $u<0.3$}\\ 0 \quad \mbox{if $u>0.3$}\\ \end{array} \right.$$It is expected that if this is repeated many times 30% $X=1$ and 70% $X=0$
u = runif(100)
x = ifelse(u<0.3, yes=1, no=0)
x
- 0
- 0
- 1
- 1
- 0
- 1
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 1
- 0
- 0
- 0
- 1
- 0
- 1
- 1
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 1
- 1
- 0
- 1
- 0
- 0
- 0
- 0
- 1
- 0
- 0
- 0
- 1
- 1
- 1
- 1
- 1
- 0
- 0
- 0
- 1
- 0
- 0
- 1
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 0
- 1
- 0
- 1
- 0
- 0
- 0
- 0
- 0
- 1
- 0
- 0
- 1
- 0
- 0
- 1
- 0
- 0
- 1
- 0
- 0
- 0
- 1
- 1
- 0
- 0
- 1
- 1
- 0
- 1
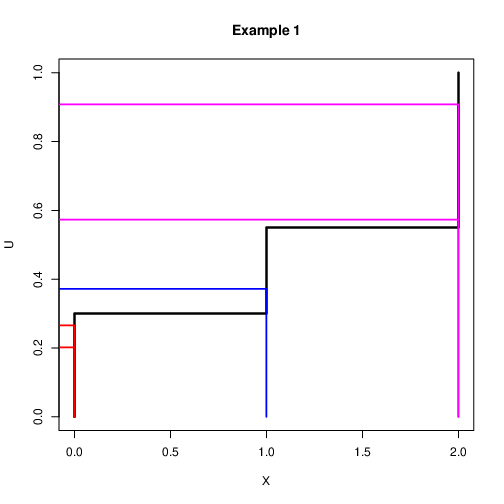
Discrete Case: Example 2¶
Assume that we want to simulate a discrete variable $X$ with probability $\mbox{Pr}(X=0)=0.3$ and $\mbox{Pr}(X=1)=0.25$ and $\mbox{Pr}(X=2)=0.45$
Let $u\sim U(0,1)$. Then the following rule can be used $$x=\left{\begin{array}{l}
0 \quad \mbox{if $u<0.3$}\\ 1 \quad \mbox{if $0.3<u<0.55$}\\ 2 \quad \mbox{if $u>0.55$} \end{array} \right.$$It is expected that if this is repeated many times, roughly 30% $X=0$, 25% $X=1$ and 45% $X=2$
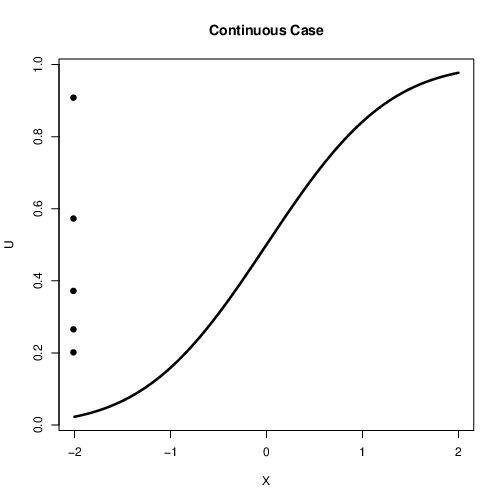
Continuous Case¶
How do we extend this idea to the continuous case?
What was the step function in our discrete example?
It is the cumulative distribution function (CDF)
Can we replace the discrete cdf with a continuous cdf?
Yes!
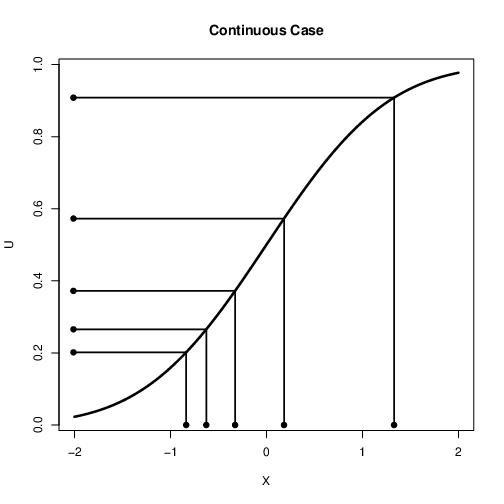
Continuous Case¶
The cdf, $F(X)$ takes values of $X$ and gives a value between $0$ and $1$
Here we take values between $0$ and $1$ and get a value of $X$
What function do we use?
We use the Inverse cdf
Continuous Case: Direct Methods¶
Probability Integral Transform¶
If $Y$ is a continuous random variable with cdf $F(y)$ , then the random variable $F_y^{-1}(U)$, where $U \sim uniform(O, 1)$, has distribution $F(y)$.
Example: If $Y \sim exponential(A)$, then the probability density function (PDF) is $$f(x;\lambda) = \begin{cases} \lambda e^{-\lambda x} & x \ge 0, \\ 0 & x < 0. \end{cases}$$ and the cumulative distribution function (CDF) is given by $$F(x;\lambda) = \begin{cases} 1-e^{-\lambda x} & x \ge 0, \\ 0 & x < 0. \end{cases}$$ Then $$F_Y^{-1}(U) = -\log(1-U)/\lambda$$ is an exponential random variable.
Thus, if we generate $U_1,..., U_n$ as iid uniform random variables, $-\lambda (1-U_i)$, are iid exponential random variables with parameter $\lambda$.
u2x <- function(u, lambda)
{
x <- - log(1-u)/lambda
return(x)
}
rmyexp <- function(n, lambda)
{
u <- runif(n, 0, 1)
x <- u2x(u, lambda)
return(x)
}
n = 1000
lambda = 2
par(mfrow = c(1, 2))
ylim <- c(0, 2)
xlim <- c(0, 4)
hist(rmyexp(n, lambda), breaks = 20, freq = FALSE,
xlim = xlim, ylim = ylim, xlab="", ylab="", main="rmyexp")
xax <- seq(0, max(x), 0.01)
xDens <- lambda*exp(-lambda*xax)
lines(xax, xDens, type = "l", col = "blue", lwd = 3)
hist(rexp(n, rate = lambda), breaks = 20, freq = FALSE,
xlim = xlim, ylim = ylim, xlab="", ylab="", main="rexp")
lines(xax, xDens, type = "l", col = "blue", lwd = 3)
Indirect Methods¶
Indirect simulation
What if the cumulative distribution function is difficult to invert, or not even available?
How to invert the CDF of a standard normal distribution?
$$F(x)=\int_{-\infty}^{x}(2\pi)^{-1/2}e^{-x^2/2}dx$$
It is still possible to simulate from this type of distribution?
If the density is available, then the answer is YES!
The mixture of normal distribution
The mixture of normal distribution has density function
$$f(x, \omega, \mu_1, \mu_2, \sigma_1, \sigma_2) = \omega N(x, \mu_1, \sigma_1) + (1- \omega) N(x, \mu_2, \sigma_2)$$
where $0 < \omega <1$ is the weight.
It is based on the combination of the normal but has many features.
It is not so easy to simulate from this distribution using the direct method.
w = 0.2
mu1 = -1
mu2 = 1
sigma1 = 0.8
sigma2 = 0.5
xgrid<-seq(-3, 4, 0.01)
nm <- dnorm(xgrid)
mix <-w*dnorm(xgrid, mu1, sigma1) + (1-w)*dnorm(xgrid, mu2, sigma2)
plot(xgrid, nm, "l", lwd=2, col='red',main='Normal: Red, Mixture: Blue',
xlab = 'x', ylab = 'y', ylim=c(0, max(nm,mix)))
lines(xgrid, mix, lwd=2,col='blue')
The idea
Let $f_y(x)$ be the target distribution and $f_v(v)$ be the proposal distribution
Simulate an $x$-coordinate from the proposal $f_v(x)$
Simulate a y-coordinate from $U(0, M*f_v(x))$
Reject any points that are not ‘inside’ $f_y(x)$
M0 = (w*dnorm(xgrid, mu1, sigma1) + (1-w)*dnorm(xgrid, mu2, sigma2))/dnorm(xgrid)
plot(xgrid, M0, type="l", xlab="x")
M = max(M0)
Mnm <-M*nm
plot(xgrid, Mnm, lwd=2, type="l",col='red',main='Scaled normal: Red, Mixture: Blue',xlab = 'x',ylab = 'y')
lines(xgrid, mix, lwd=2, col='blue')
Reject and accept method¶
Theorem: Let $Y \sim f_Y(y)$ and $V \sim f_v(v)$, where $f_Y$ and $f_V$ have common support with $$M = sup_y f_Y(y)/f_V(y) < \infty.$$ To generate a random variable $Y\sim f_Y$, we do the following steps
Generate $U\sim uniform(0,1)$ and $V\sim f_V$ independently.
If $$U < \frac{1}{M} \frac{f_Y(V)}{f_V(V)}$$ set $Y = V$; otherwise, return to step 1.
n<-500
xp<-rnorm(n)
yp<-M*dnorm(xp)*runif(n)
plot(xgrid, Mnm, lwd=2, type="l",col='red',main='Scaled Normal: Red, Mixture: Blue',xlab = 'x', ylab = 'y')
lines(xgrid, mix, lwd=2, col='blue')
dmix = w*dnorm(xp, mu1, sigma1) + (1-w)*dnorm(xp, mu2, sigma2)
points(xp[yp>=dmix],yp[yp>=dmix], col='red',pch=20, cex=0.2)
points(xp[yp<dmix],yp[yp<dmix], pch=16, cex=0.2, col='blue')
Reject and accept method: the proof¶
$$\begin{aligned} P(V\leq y |stopping~rule)&= P(V\leq y | U < \frac{1}{M} \frac{f_Y(V)}{f_V(V)})\\ &= \frac{P(V\leq y , U < \frac{1}{M} \frac{f_Y(V)}{f_V(V)})}{P(U < \frac{1}{M} \frac{f_Y(V)}{f_V(V)})}\\ &= \frac{\int_{-\infty}^y\int_0^{\frac{1}{M} \frac{f_Y(V)}{f_V(V)}}du~f_V(v)dv}{\int_{-\infty}^{\infty}\int_0^{\frac{1}{M} \frac{f_Y(V)}{f_V(V)}}du~f_V(v)dv}\\ &= \frac{\int_{-\infty}^y\frac{1}{M} \frac{f_Y(V)}{f_V(V)}f_V(V)dv}{\int_{-\infty}^{\infty}\frac{1}{M} \frac{f_Y(V)}{f_V(V)}f_V(V)dv}\\ &=\int_{-\infty}^y f_Y(v)dv \end{aligned}$$- Can have any $M$? In fact $M$ shows the efficiency of the sampling algorithm, i.e. $M= 1/P(stopping~rule)$.
Choosing $f_v(x)$ and $M$¶
Two things are necessary for the accept/reject algorithm to work:
The domain of $p(x)$ and the domain of $q(x)$ MUST be the same.
The value of $M$ must satisfy $M\geq \sup_x p(x)/q(x)$
The algorithm will be more efficient if:
The proposal $q(x)$ is a good approximation to $p(x)$
The value of $M$ is $M=\sup_x p(x)/q(x)$
A look at $f_y(x)/f_v(x)$¶
Normalizing Constant and Kernel
Today we saw many density functions $f(x)$
Many density functions can written as $f(x)=k\tilde{f}(x)$
The part $k$ is called the normalizing constant and the part $\tilde{f}(x)$ is called the kernel.
For example the standard normal distribution is $$p(x)=(2\pi)^{-1/2}e^{-x^2/2}$$
Normalizing Constant and Kernel What are the normalizing constant and kernel of the Beta function?
$$Beta(x;a,b)=\frac{\Gamma(a+b)}{(\Gamma(a)\Gamma(b))}{x^{a-1}(1-x)^{b-1}}$$Accept/Reject and the normalising constant¶
An advantage of the Accept/Reject algorithm is that is works, even if the normalizing constant is unavailable.
Only the kernel is needed.
There are many examples where the normalising constant is either unavailable or difficult to compute.
This often happens in Bayesian analysis